-Search query
-Search result
Showing all 27 items for (author: kuhle & b)

EMDB-29070: 
Cryo-electron microscopy structure of human mt-SerRS in complex with mt-tRNA(UGA-TL)
Method: single particle / : Hirschi M, Kuhle B, Doerfel L, Schimmel P, Lander G

PDB-8ffy: 
Cryo-electron microscopy structure of human mt-SerRS in complex with mt-tRNA(UGA-TL)
Method: single particle / : Hirschi M, Kuhle B, Doerfel L, Schimmel P, Lander G
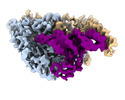
EMDB-26310: 
Cryo-electron microscopy structure of human mt-SerRS in complex with mt-tRNA (GCU)
Method: single particle / : Hirschi M, Kuhle B, Doerfel L, Schimmel P, Lander G
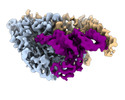
EMDB-26311: 
Cryo-electron microscopy structure of human mt-SerRS in complex with mt-tRNA(GCU-TL)
Method: single particle / : Hirschi M, Kuhle B, Doerfel L, Schimmel P, Lander G

PDB-7u2a: 
Cryo-electron microscopy structure of human mt-SerRS in complex with mt-tRNA (GCU)
Method: single particle / : Hirschi M, Kuhle B, Doerfel L, Schimmel P, Lander G

PDB-7u2b: 
Cryo-electron microscopy structure of human mt-SerRS in complex with mt-tRNA(GCU-TL)
Method: single particle / : Hirschi M, Kuhle B, Doerfel L, Schimmel P, Lander G

EMDB-10495: 
Cryo-EM Structure of NADH reduced form of NAD+-dependent Formate Dehydrogenase from Rhodobacter capsulatus
Method: single particle / : Wendler P, Radon C, Mittelstaedt G

EMDB-10496: 
Cryo-EM Structure of as isolated form of NAD+-dependent Formate Dehydrogenase from Rhodobacter capsulatus
Method: single particle / : Wendler P, Radon C, Mittelstaedt G

EMDB-10093: 
Structure of the core of the flagellar export apparatus from Vibrio mimicus, the FliPQR-FlhB complex.
Method: single particle / : Kuhlen L, Johnson S, Deme JC, Lea SM

EMDB-10095: 
Structure of the FliPQR complex from the flagellar type 3 secretion system of Pseudomonas savastanoi.
Method: single particle / : Kuhlen L, Johnson S, Deme JC, Lea SM

EMDB-10096: 
Structure of the FliPQR complex from the flagellar type 3 secretion system of Vibrio mimicus.
Method: single particle / : Kuhlen L, Johnson S, Deme JC, Lea SM

EMDB-10653: 
Structure of the core of the flagellar export apparatus from Vibrio mimicus, the FliPQR-FlhB, in amphipol A8-35
Method: single particle / : Kuhlen L, Johnson S, Deme JC, Lea SM

PDB-6s3l: 
Structure of the core of the flagellar export apparatus from Vibrio mimicus, the FliPQR-FlhB complex.
Method: single particle / : Kuhlen L, Johnson S, Deme JC, Lea SM

PDB-6s3r: 
Structure of the FliPQR complex from the flagellar type 3 secretion system of Pseudomonas savastanoi.
Method: single particle / : Kuhlen L, Johnson S, Deme JC, Lea SM

PDB-6s3s: 
Structure of the FliPQR complex from the flagellar type 3 secretion system of Vibrio mimicus.
Method: single particle / : Kuhlen L, Johnson S, Deme JC, Lea SM

EMDB-4733: 
Improved map of the FliPQR complex that forms the core of the Salmonella type III secretion system export apparatus.
Method: single particle / : Johnson S, Kuhlen L, Abrusci P, Lea SM

EMDB-4734: 
Structure of the core Shigella flexneri type III secretion system export gate complex SctRST (Spa24/Spa9/Spa29).
Method: single particle / : Johnson S, Kuhlen L, Deme JC, Abrusci P, Lea SM

PDB-6r69: 
Improved map of the FliPQR complex that forms the core of the Salmonella type III secretion system export apparatus.
Method: single particle / : Johnson S, Kuhlen L, Abrusci P, Lea SM

PDB-6r6b: 
Structure of the core Shigella flexneri type III secretion system export gate complex SctRST (Spa24/Spa9/Spa29).
Method: single particle / : Johnson S, Kuhlen L, Deme JC, Abrusci P, Lea SM

EMDB-0149: 
PTC3 holotoxin complex from Photorhabdus luminecens in prepore state (TcdA1, TcdB2, TccC3)
Method: single particle / : Gatsogiannis C, Merino F, Roderer D, Balchin D, Schubert E, Kuhlee A, Hayer-Hartl M, Raunser S

EMDB-0150: 
PTC3 holotoxin complex from Photorhabdus luminiscens - Mutant TcC-D651A
Method: single particle / : Gatsogiannis C, Merino F, Roderer D, Balchin D, Schubert E, Kuhlee A, Hayer-Hartl M, Raunser S

PDB-6h6e: 
PTC3 holotoxin complex from Photorhabdus luminecens in prepore state (TcdA1, TcdB2, TccC3)
Method: single particle / : Gatsogiannis C, Merino F, Roderer D, Balchin D, Schubert E, Kuhlee A, Hayer-Hartl M, Raunser S

PDB-6h6f: 
PTC3 holotoxin complex from Photorhabdus luminiscens - Mutant TcC-D651A
Method: single particle / : Gatsogiannis C, Merino F, Roderer D, Balchin D, Schubert E, Kuhlee A, Hayer-Hartl M, Raunser S

EMDB-4173: 
A FliPQR complex forms the core of the Salmonella type III secretion system export apparatus.
Method: single particle / : Johnson S, Kuhlen L, Abrusci P, Lea SM

PDB-6f2d: 
A FliPQR complex forms the core of the Salmonella type III secretion system export apparatus.
Method: single particle / : Johnson S, Kuhlen L, Abrusci P, Lea SM
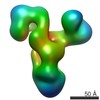
EMDB-3712: 
Human BBsome core complex with subunits BBS1, BBS4, BBS5, BBS8, BBS9, BBS18
Method: single particle / : Klink BU, Zent E, Juneja P, Kuhlee A, Raunser S, Wittinghofer A

EMDB-2280: 
Negative stain EM structure of the HOPS tethering complex (conformation 1)
Method: single particle / : Broecker C, Kuhlee A, Gatsogiannis C, Balderhaar HJ, Hoenscher C, Engelbrecht-Vandre S, Ungermann C, Raunser S
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
